research
We aspire to understand and manipulate metabolism at the systems level - broadly across the metabolic network and quantitatively considering how reactions work together - with the ultimate goal of reprogramming life from its chemistry.
Understanding metabolism at the systems level
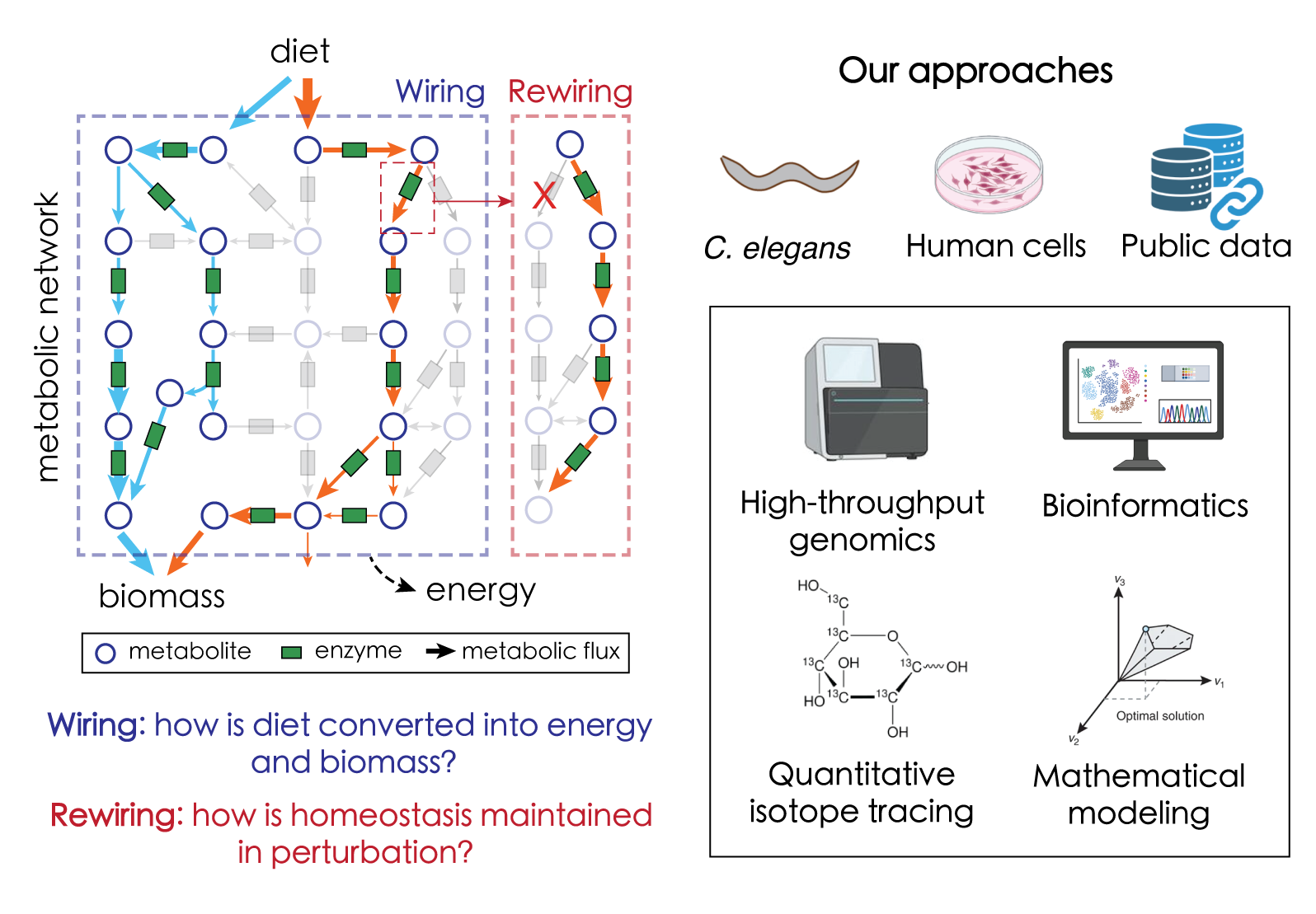
“More is Different,” a concept introduced in the field of condensed matter physics, argues that complex systems exhibit behaviors that are not simply a sum of their individual parts, but are fundamentally different and require new concepts to understand them. Metabolism is exactly the case. Driven by an intricate network of biochemical reactions, metabolism convert nutrients into energy and biomass to sustain life. No single reaction can carry out its function in isolation. Gaining a systems-level understanding of metabolism is a crucial step beyond conventional deconstructive research – which breaks metabolism down into individual biochemical reactions – toward ultimately reconstructing how these conversions of biological molecules integrate to create life.
We combine computational modeling and high-throughput experiments to dissect metabolism at the systems level. In our previous studies, we have established a novel genomic framework (instead of conventional biochemistry!) to achieve the first systems-level characterization of both function (wiring) and regulation (rewiring) of metabolism. We are expanding this paradigm-shifting approach to understand the systems-level principles of metabolism from single cells to complex multicellular organisms.
Manipulating metabolism at the systems level
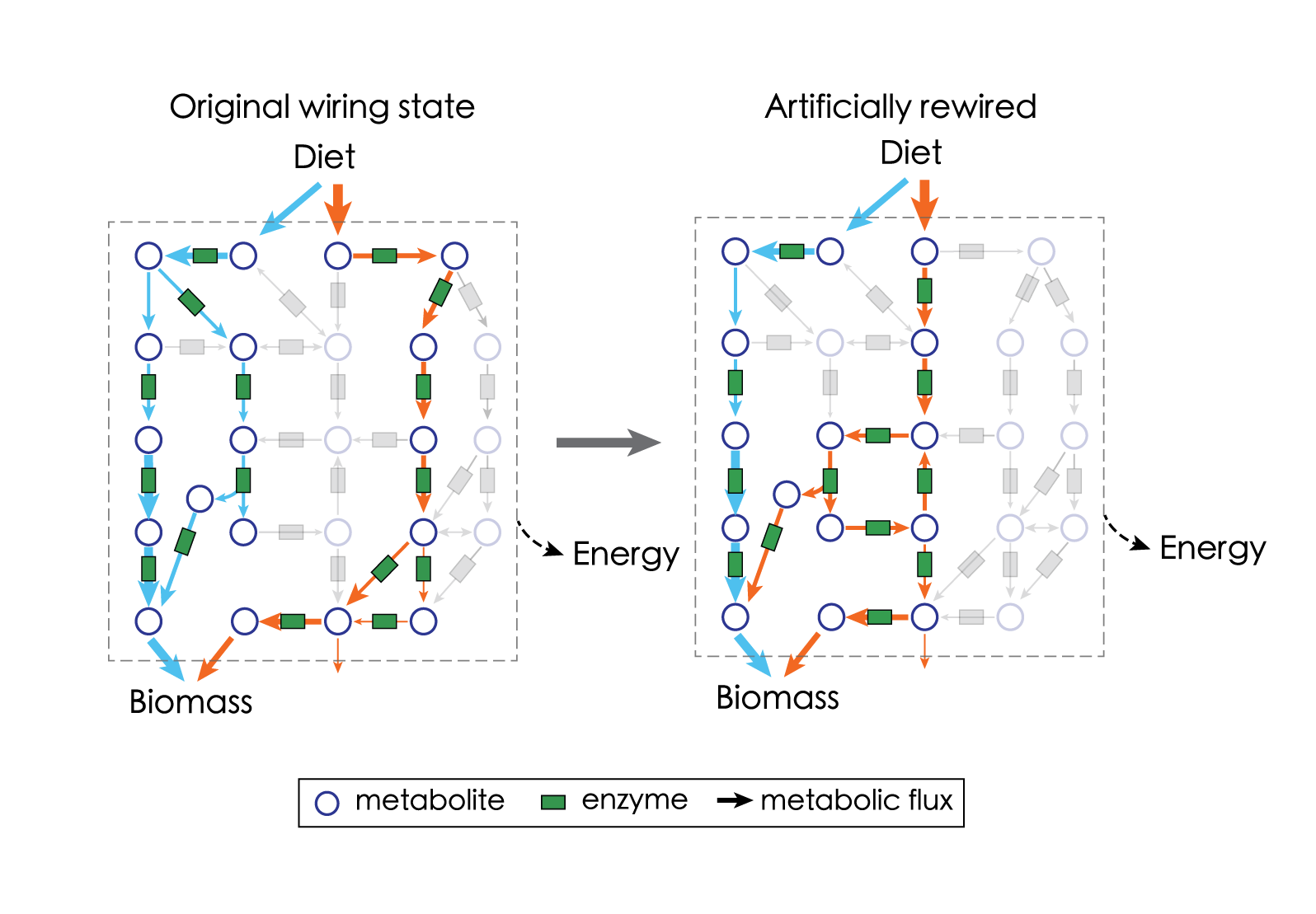
The famous physicist Richard Feynman once said, “What I cannot create, I do not understand.” The ultimate test of how well we understand metabolism is to create it. Imagine when we have learned quite a bit about how metabolism runs in normal cells and how it is regulated in response to perturbations – what can we do next? An exciting direction will be to apply the principles we’ve learned to guide the manipulation of metabolism.
We are actively exploring both computational and experimental approaches to artificially ‘rewire’ and even ‘wire’, metabolism at the systems level. This is often referred to as metabolic engineering – but we have something new! We are integrating cutting-edge approaches such as AI-based protein design to gain the systems-level control of metabolism. Our goal is to translate the principles we understand about metabolism into what we could create.
A systems solution to errors in metabolism
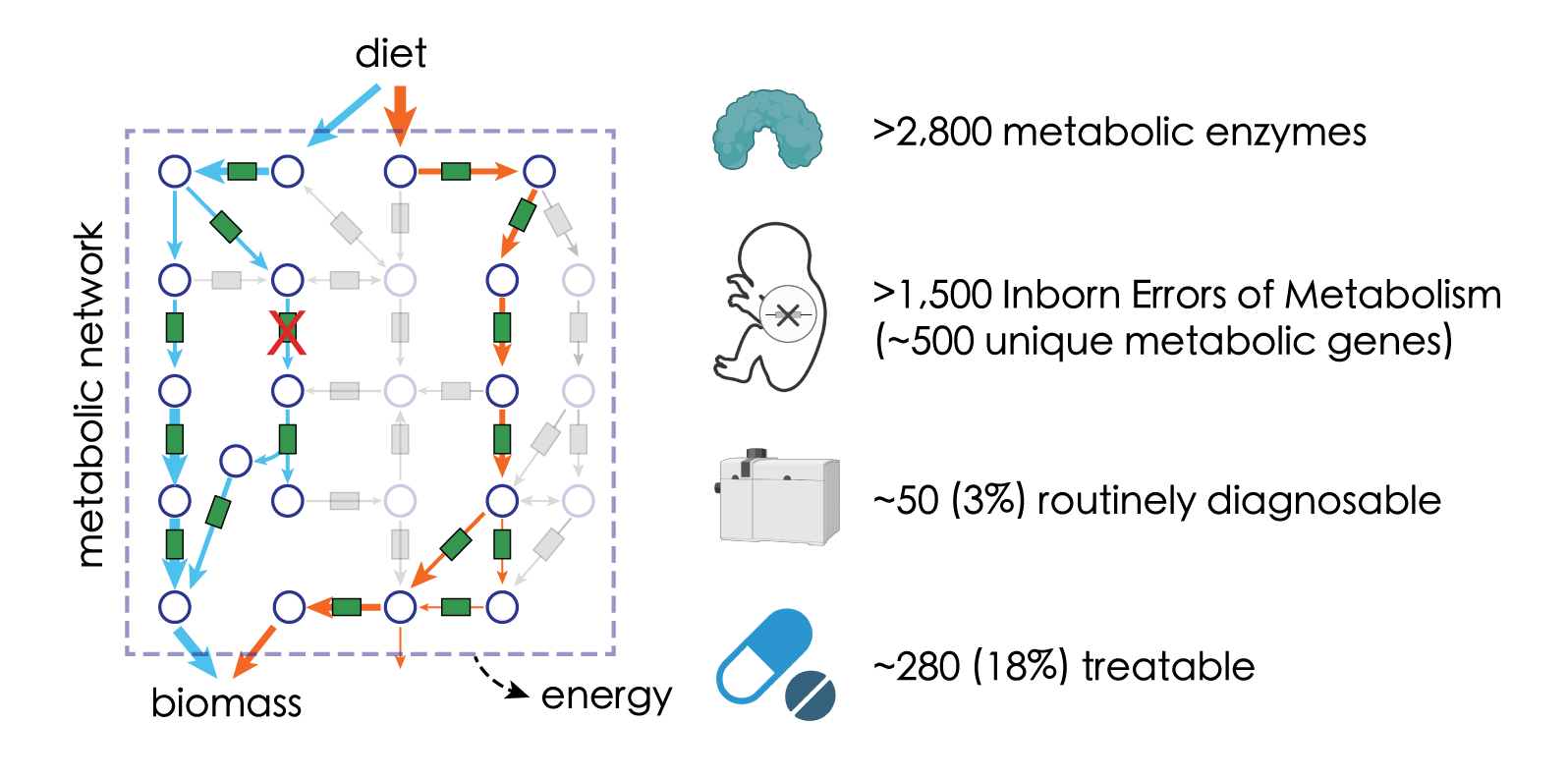
Like many other processes in life, metabolism can cause diseases when it goes wrong. Diseases caused by mutations in metabolic genes (enzymes) are called Inborn Errors of Metabolism (IEM). IEM is a class of pediatric diseases that includes over 1,500 individual cases, covering errors in almost every parts of metabolism – carbohydrates, amino acids, fatty acids, etc. Despite that most IEMs are (ultra) rare diseases, they collectively affect every 1 in 2,000 newborns worldwide. Devastatingly, most IEMs cannot be effectively treated – nor even timely diagnosed – as most of them are rare diseases that are not often screened in clinic and rarely studied in research.
We seek to translate our research in metabolic systems biology into clinical benefits for diagnosing and treating IEMs. For instance, our systematic study of metabolic gene perturbations may reveal common characteristics among various IEMs to guide “one-for-many” therapeutic interventions. Integrating these big data with AI, our goal is to develop novel diagnostic and therapeutic approaches that can address many IEMs at once.